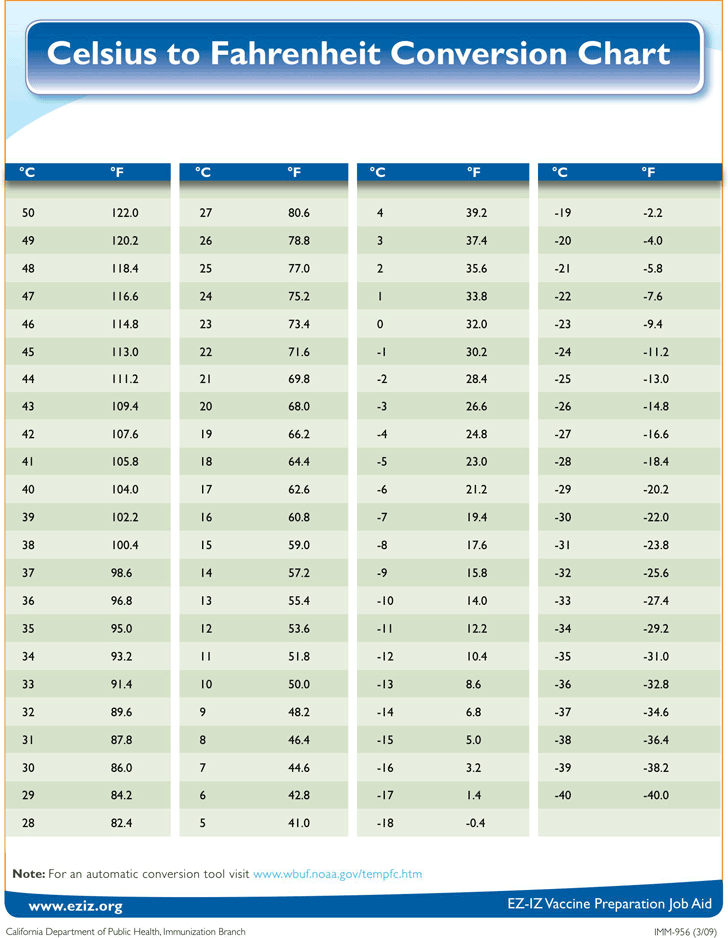
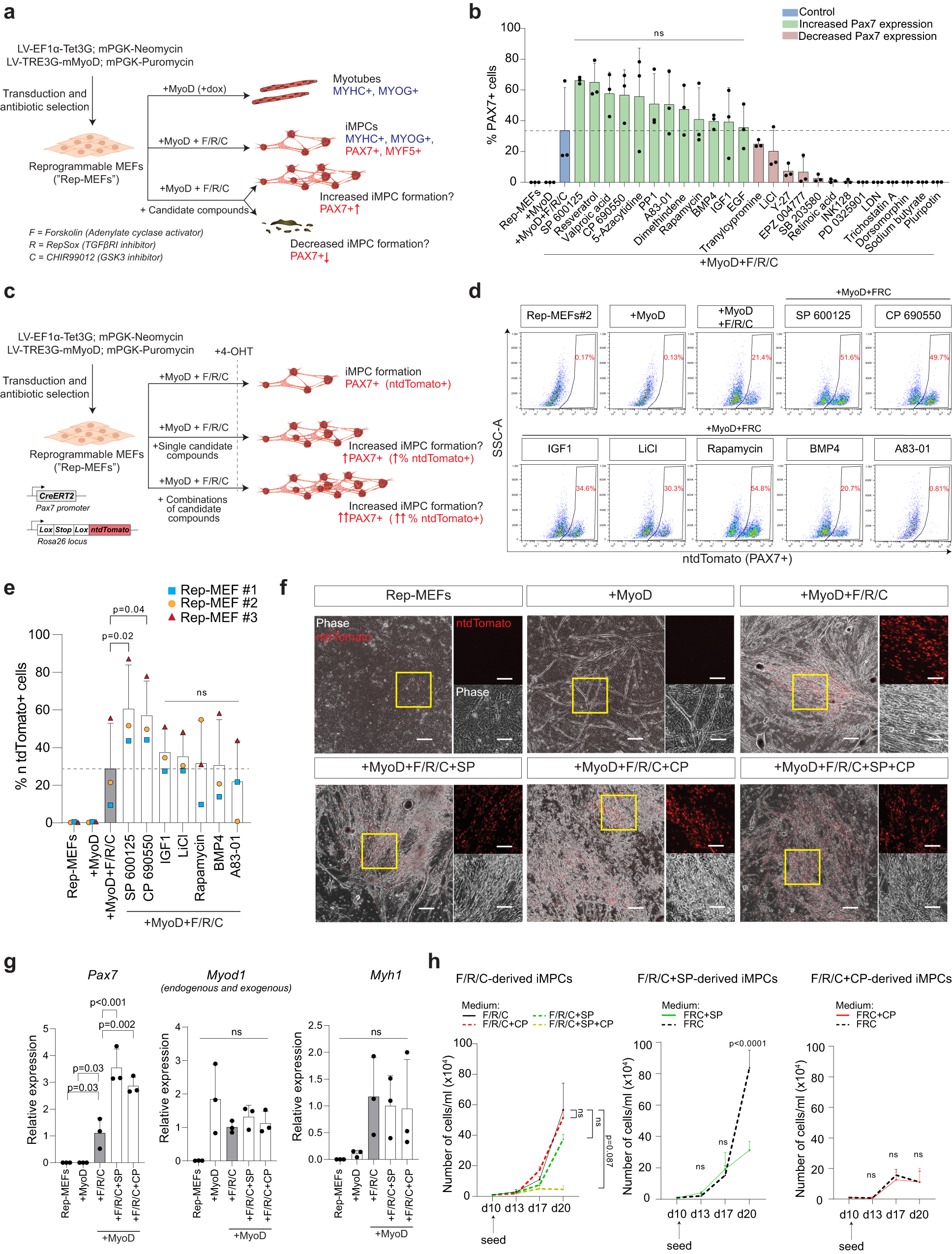
dataset - Is there a way to convert smiles format to TUdataset
5 (630) · $ 33.00 · In stock
How to convert the molecular graph in SDF/JSON format to the input of graphormer? · Issue #108 · microsoft/Graphormer · GitHub
What are some ways to present a large dataset in a visually appealing manner? - Quora
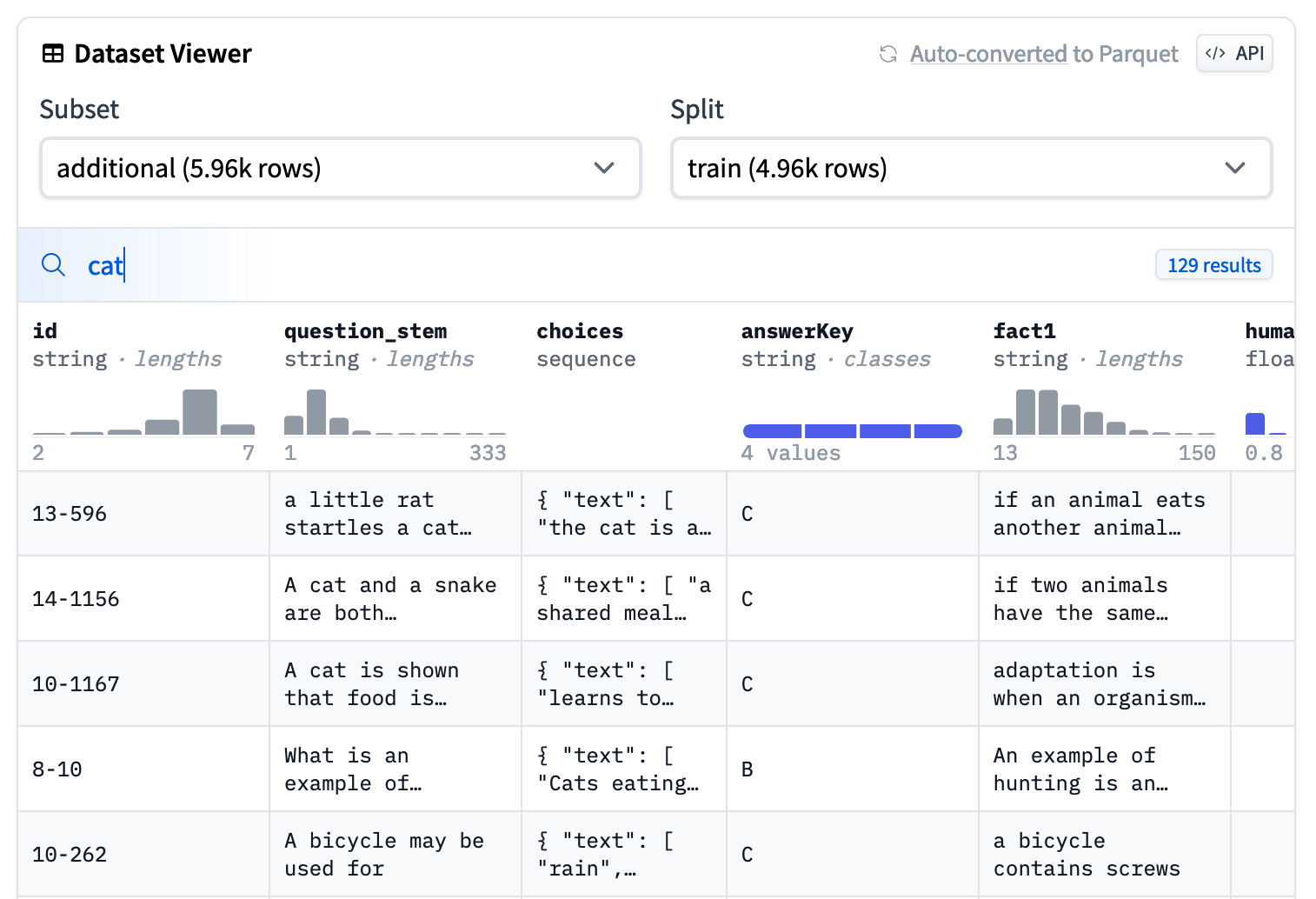
Dataset viewer

How to a Bundle of .smiles convert into .sdf file format

Relating the xyz files to the molecules in PCQM4Mv2 · Issue #297 · snap-stanford/ogb · GitHub
sdftosmi.py: Convert multiple ligands/compounds in SDF format to SMILES. — Bioinformatics Review

sdftosmi.py: Convert multiple ligands/compounds in SDF format to SMILES. — Bioinformatics Review
www.
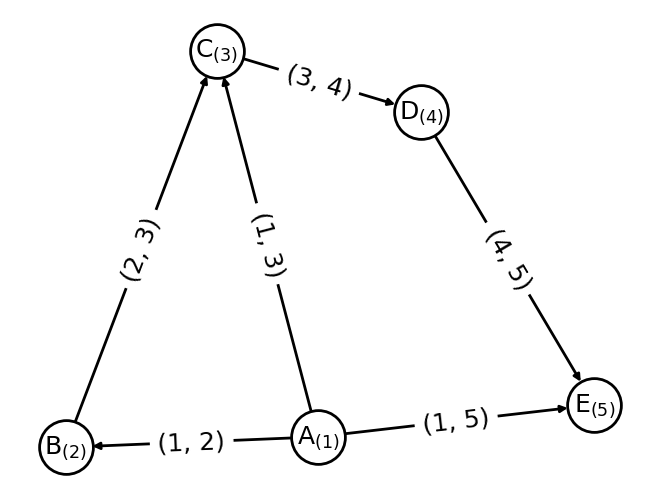
Data — kgcnn 2.2.1 documentation

dataset - Is there a way to convert smiles format to TUdataset format? - Stack Overflow
www.
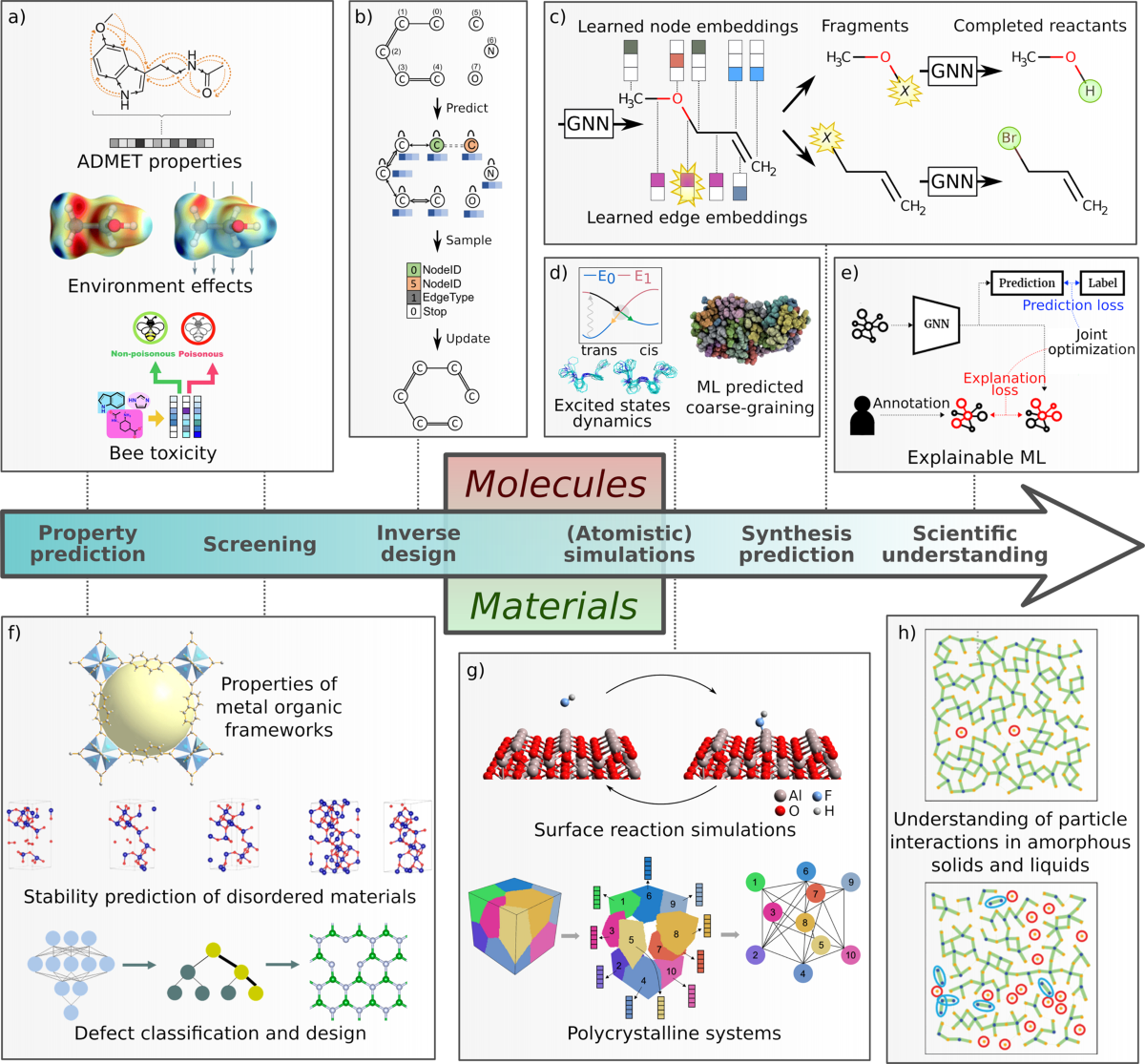
Graph neural networks for materials science and chemistry
GitHub - RohanV01/Molecule_format_converter: This jupyter notebook let's you convert sdf to pdb or SMILES to pdbqt file formats for a batch of compounds to perform cheminformatics and drug discovery projects
gcnn_keras/docs/source/data.ipynb at master · aimat-lab/gcnn_keras · GitHub